Building High-quality Knowledge Graphs. Using and developing Knowledge Graph technologies and methods to structured data and to connect them to existing biological knowledge. These structures facilitate analysis and interpretation of complex data. We are contributing to a groundbreaking field by developing tools and methods to build, assess and investigate Knowledge Graphs and applying them to solve challenges in biology and health.
In this project, we investigated the microbiome of the wastewater treatment (WWT) process to build MicW2Graph, an open-source knowledge graph that integrates metagenomic and metatranscriptomic information with their biological context, including biological processes, environmental and phenotypic features, chemical compounds, and additional metadata. We developed a workflow to collect meta-omics datasets from MGnify and infer potential interactions among microorganisms through microbial association networks. MicW2Graph enables the investigation of research questions related to WWT, focusing on aspects such as microbial connections, community memberships, and potential ecological functions.
The following figure shows the general workflow of the MicW2Graph project:

KGBioGraphy is a manually-curated knowledge graph that contains information of the data sources and usecases of published biological/biomedical knowledge graphs (BKGs). Currently, there are 69 BKGs summarized within KGBioGraphy. Each BKG within KGBioGraphy is represented by an ego network, linking the BKG to its publication, data sources (e.g., databases and ontologies), node and relationship types it contains, and usecases.
We incorporated the open access BKG publications and KGBioGraphy into a corrective Retrievcal-Augmented Generation (RAG) model which provides a large language model (LLM) with a context-rich prompt to improve LLM response performance. Coming soon: Users will soon be able to interact with the KG-BioGraphy RAG through a streamlit interface and API.
The following is a flow diagram of the Retrieval-Augmented Generation (RAG) model and KG-BioGraphy:
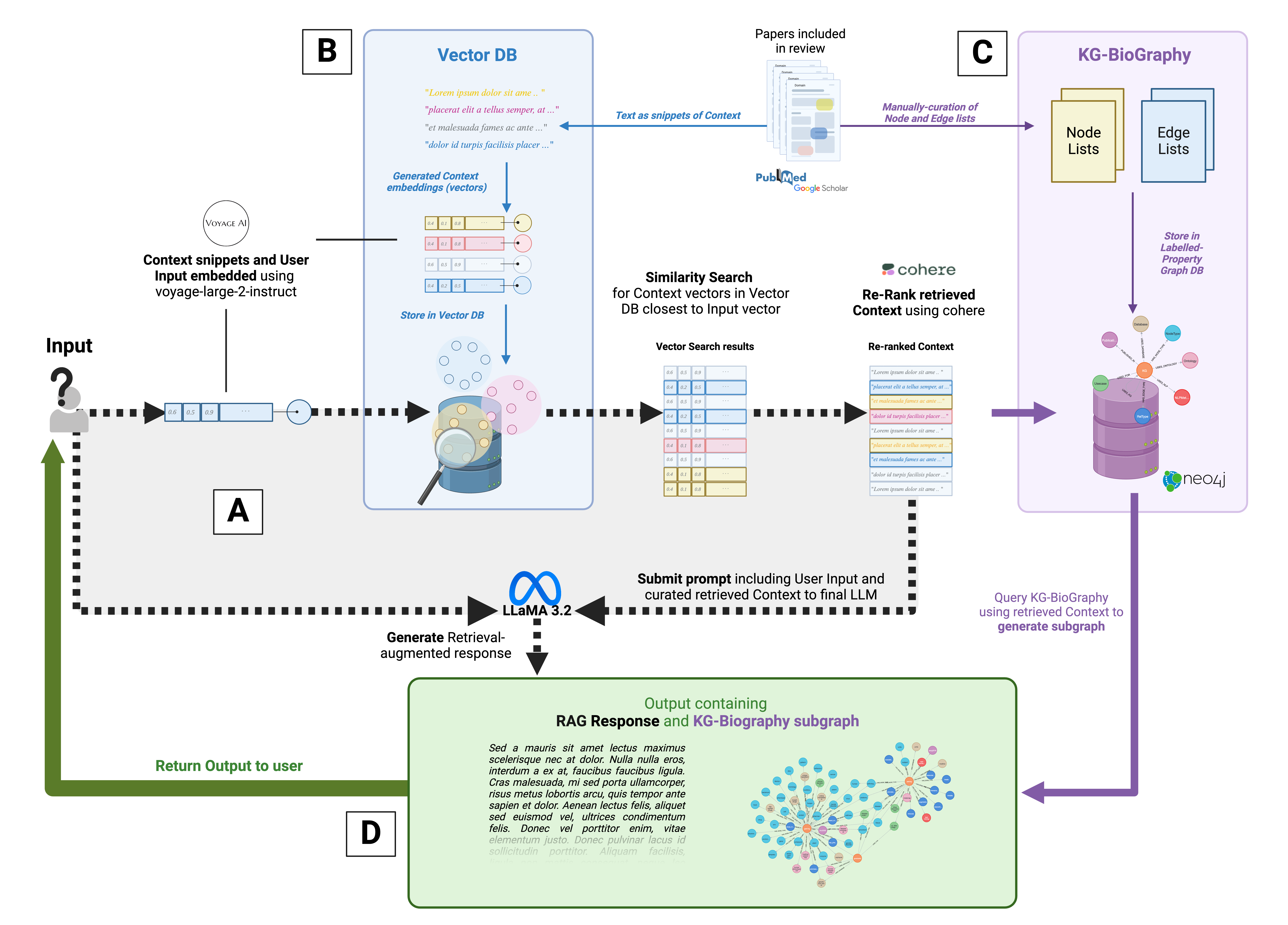
(A) RAG Architecture workflow.
(B) A vector database (DB) comprising vector representations of text snippets from the publications included in the review.
(C) KG-BioGraphy Neo4j DB.
(D) The retrieved contexts are used to query 1) the LLM and 2) the KG-BioGraphy DB to generate a text and subgraph response, respectively, which is outputed to the user.
This figure was created on Biorender.com.
Developing and Applying Novel Methods on Graphs. Unleashing the power of Machine Learning on Graphs, a cutting-edge approach to extracting valuable insights from network data. We explore how this fusion of machine learning and graph theory helps to recognize patterns, generate predictions, and discovering new knowledge across a multitude of applications, including biological and medical networks.
Exploring Microbial Communities and their Environments. Integrating multiple biological resources to unravel the assembly, interaction and adaptation mechanisms of microbial networks, offering insights into their functions and inpact on ecosystems, and how changes affect those communities.
Implementing tools to process, integrate, and analyse multimodal data. Diving into the benefits of harmonising multimodal data that converge to provide a comprehensive view of complex biological systems. Specifically we are interested in high-throughput multi-omics data generated using Mass spectrometry technology (proteomics and metabolomics) and metaomics data (metagenomics and metaproteomics).
Data Science Democratisation. Focusing on data literacy training as a means to reduce inequality, and promoting open science by making all research, data content, and software open and accessible.
Ayala-Ruano, S., Webel, H., & Santos, A. (2025). VueGen: Automating the generation of scientific reports. bioRxiv. https://doi.org/10.1101/2025.03.05.641152