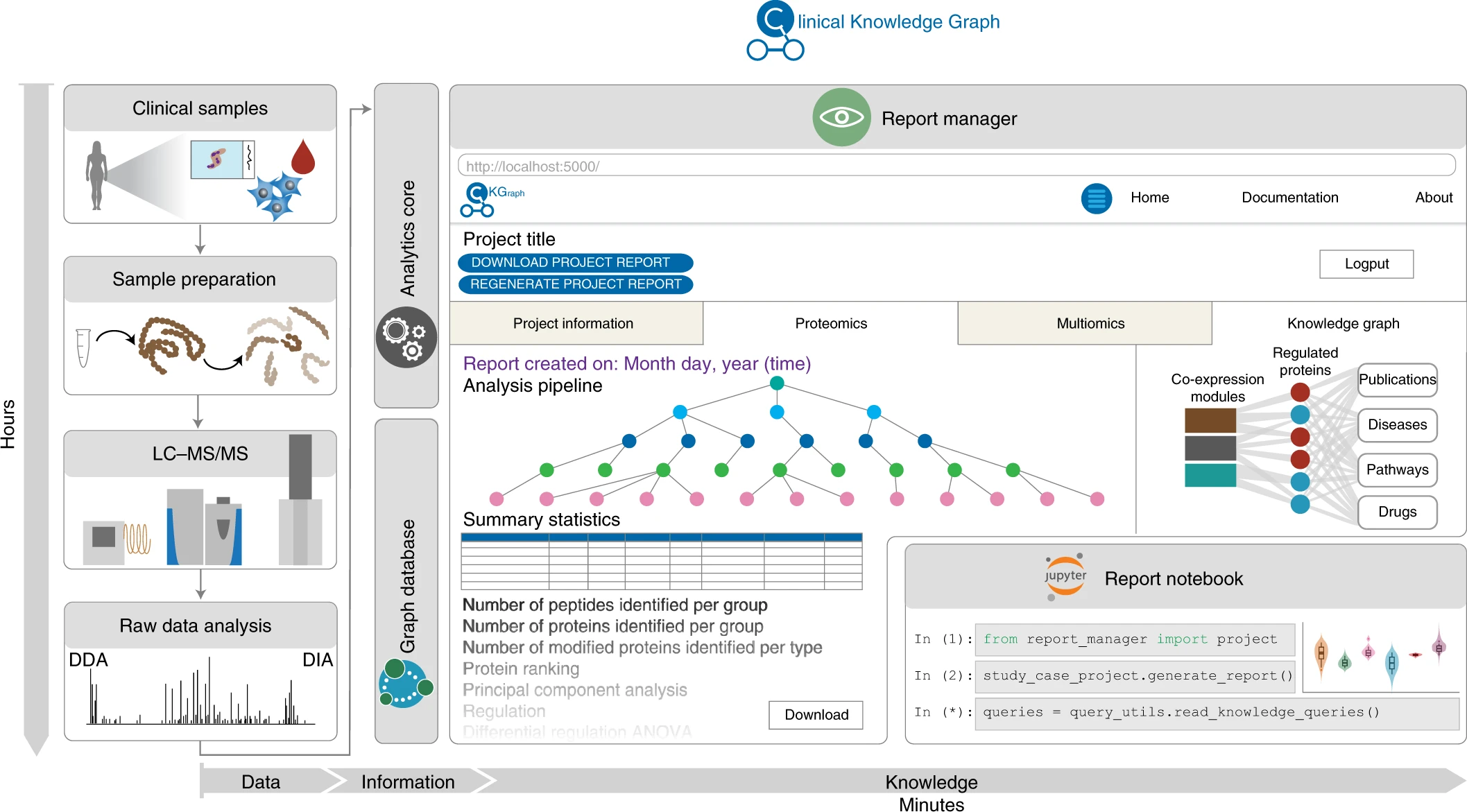
The Clinical Knowledge Graph (CKG) is a graph database with millions of nodes and relationships representing experimental data, databases, and scientific literature. Further, CKG is an open-source platform that allows easy expansion with new databases and experimental data types and it includes statistical and machine learning tools for faster proteomics analysis.
We are currently working on a new version that will be more flexible and generalizable. Stay tuned!
Understanding host-parasite interactions is vital for tackling infectious diseases. Due to the challenges in experimental identification, we propose a computational method to predict protein interactions between human and 18 eukaryotic parasites. Our approach leverages an orthology-based transfer of interactions, focusing on parasite secretomes and human membrane proteins. We also filter the host proteome based on the parasites' specific tissue tropisms. In this version, we added cell-type specific expression annotations to provide further resolution of the host-parasite predicted interactions and we support interactions with structural information.
Acore, short for analytics core, is an open-source Python library to preprocess and analyse multi-omics data. It includes functionality related to preprocessing, e.g. for data normalization, missing value imputation or feature selection, and functionality for statistical data analysis, e.g. an analysis of covariance.
Acore is designed to be user-friendly and flexible, allowing users to easily apply different analysis strategies, testing effects of choosing specific steps.
VueGen is a tool that automates the creation of reports from bioinformatics outputs, allowing researchers with minimal coding experience to communicate their results effectively. With VueGen, users can produce reports by simply specifying a directory containing output files, such as plots, tables, networks, Markdown text, HTML components, and API calls, along with the report format. Supported formats include documents (PDF, HTML, DOCX, ODT), presentations (PPTX, Reveal.js), Jupyter notebooks and Streamlit web applications.
An overview of the VueGen workflow is shown in the figure below:
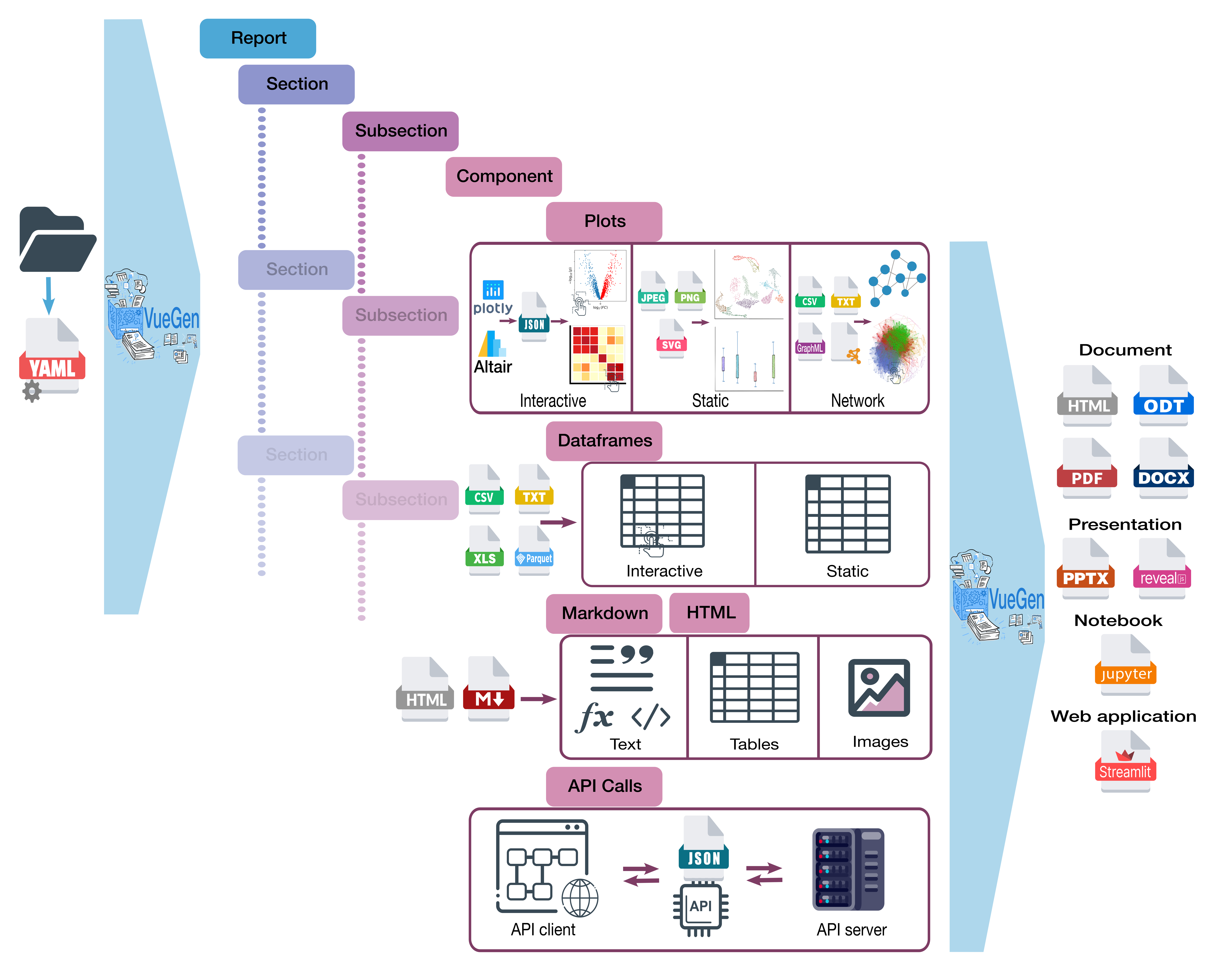
VueGen offers various implementation options for both non-technical and experienced users. It is available as a Python package, Docker image, nf-core module, and cross-platform desktop application with a user-friendly interface, making it accessible and customizable for different user needs and expertise levels.
The documentation is available at vuegen.readthedocs.io, where you can find detailed information of the package’s classes and functions, installation and execution instructions, and case studies to demonstrate its functionality.
A graphical image of FermentDB workflow is shown in the figure below:
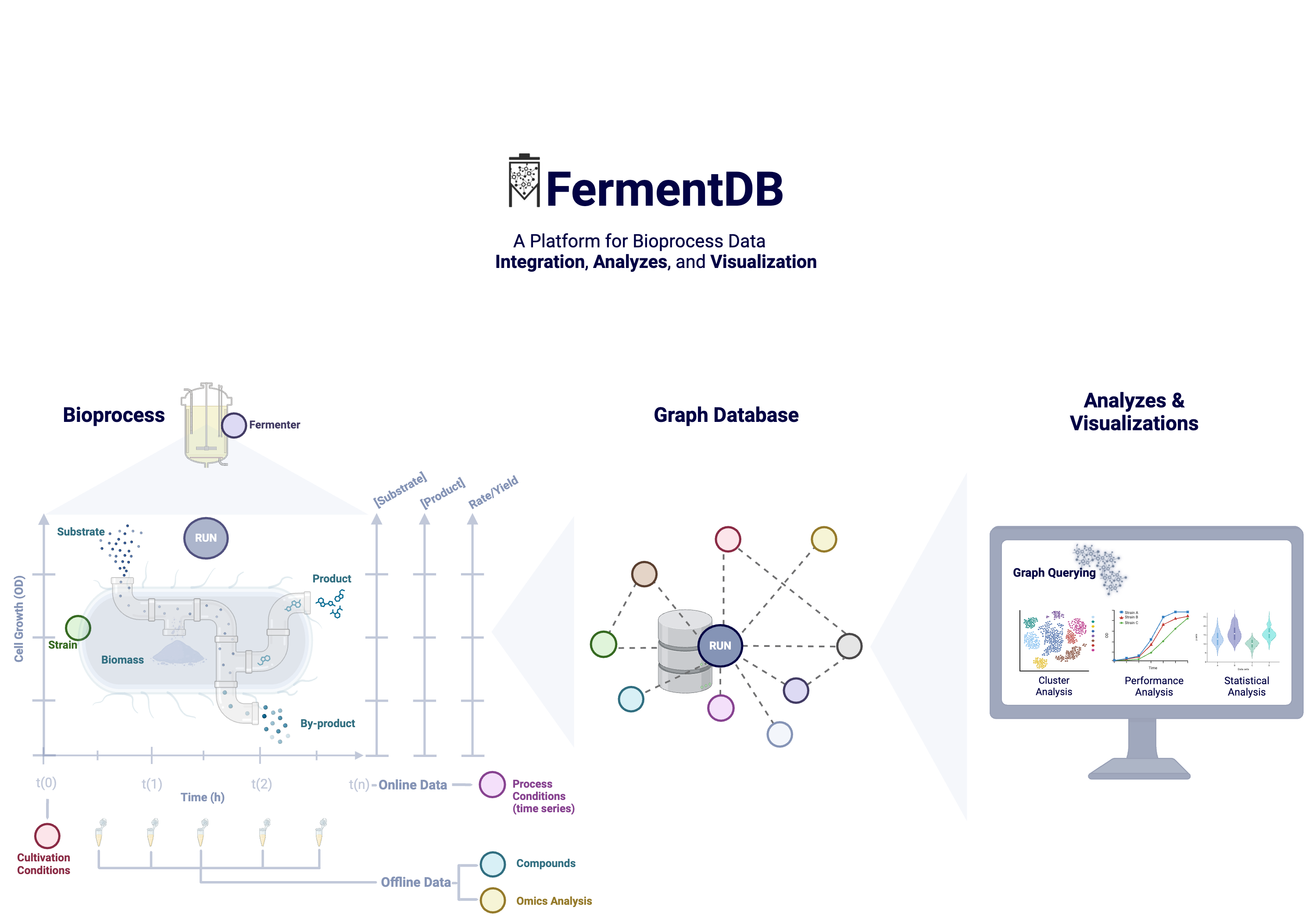
FermentDB is freely available on the websitefermentdb.streamlit.app
The application code is publicly available in GitGub: FermentDB Graph and FermentDB API.
gdbcore is a python library that can take your tables and transform them into graph databases in Networkx, graph-tool or Neo4j (v4). gdbcore is available for installation from PyPI.
We are currently improving the project and the documentation. Stay tuned!
Paragraph of text inside
More text here if you want